The app lets you build a network step by step. Nodes and edges can be added to a network in a piecemeal fashion. Nodes can represent various biological entities, such as: a pathway, a protein, a reaction, or a compound. Edges can represent any type of relation between those entities.
Here is an example workflow.
-
Search for an entity to seed your network. The General SPARQL search menu gives you a range of options. Every menu item is backed by a SPARQL query.
-
In this example we search for the HGNC symbol TPI1
-
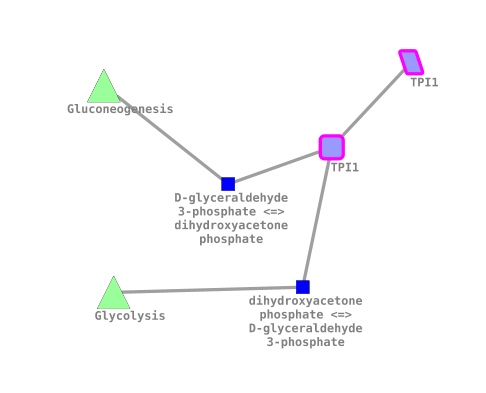
A single node is placed on the network

-
Right click on the node to open the context menu. Also this menu is backed by SPARQL queries. We can find the associated protein for this gene.

-
By right-clicking again we can find associated reactions and pathways from reactome
-
After expanding the Glycolysis node in a similar way, we can see all reactions, proteins and metabolites that are part of it. With a few more clicks we could find all related GO terms or chemical assays from Chembl.
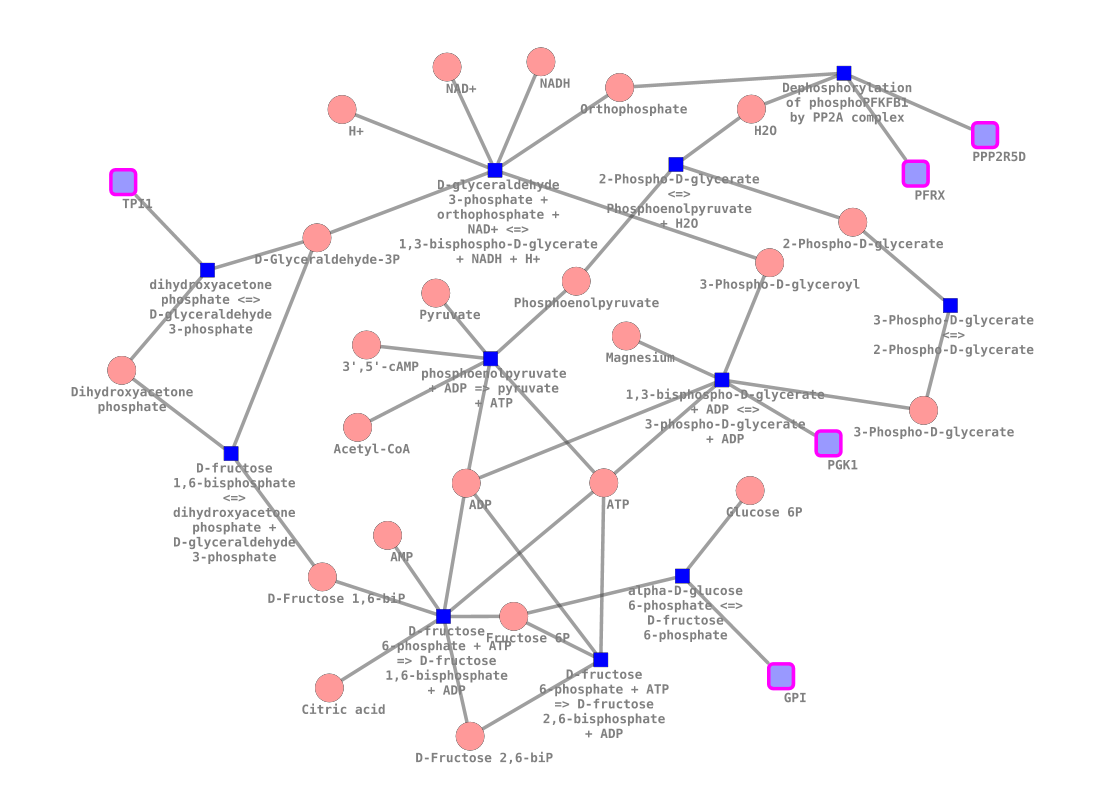
Generally speaking, you can then right-click on nodes to pull in related entities. For example, all the pathways that are related to your protein. Or all the Gene Ontology annotations. Or all the reactions that your protein is part of. Or the gene that encodes for your protein. And you can continue this process, jumping from one entity to the next.
Up: Home Previous: Installation Next: Developer Manual